If you ever want to work from home, connecting remotely to your computer is a must. In order for things to work the way you expect, you need to keep a few things in mind.
SSH
X Forwarding
- To connect to your office desktop (MSI and College of Science and Engineering Labs also offer SSH connection to their Linux machines), you will need an SSH client on your home computer or laptop. If you are working from a Mac or a Linux machine, you already have one built in. Just open a terminal and go. If you are working from a Windows PC, you can use MobaXterm (with X11 server and support for RDP, VNC, SSH, telnet, rsh, FTP, SFTP and XDMCP) or PuTTY.
- To run your X11 programs, you need some kind of X-emulator to mimic the X Window System. If you work from a Mac or a Linux machine at home, you already have one built in. If you work from a Windows PC and the client program you are using does not have it bundled, then you need VcXsrv or Xming.
- Be mindful of the firewall. Your desktop is set to deny access to any IP address coming from off-campus computers. To get around this, you can either first connect to one of MSI’s machines, and then connect to your computer, or to allow your home IP address Configure the tcp wrapper.
- To avoid all the trouble of getting SSH and X11 forwarding, as well as other Unix stuff, to work in Microsoft Windows, without the need for dual-booting, you can use Windows Subsystem for Linux, cygwin, or install a Linux distribution within your Windows operating system using VirtualBox.
- If you just want to access the library, then either VPN or SSH Tunneling should be sufficient.
SSH Tunneling
You can set up machines to which you have SSH access as Internet proxy via SSH Tunneling. This is useful for journal/database services which are restricted to university IP addresses. 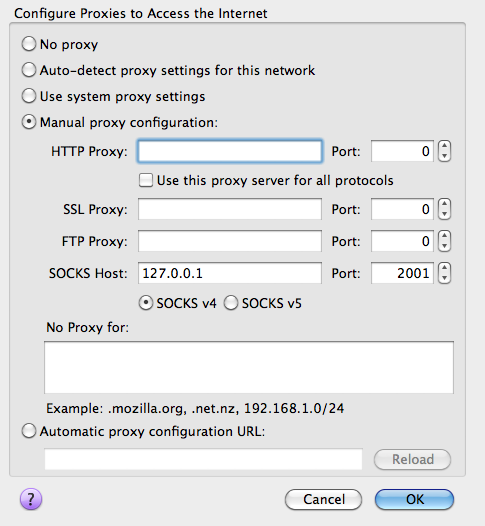
- Establish a SSH connection with dynamical port forwarding: under Linux or Mac OS X, just open a terminal and enter:
ssh -D 2001 -fN your_user_name@your_machine.dept.univ.edu. If you use PuTTY on Windows, load a saved session or create a new one, then in the left panel of the Configuration window, under Connection -> SSH -> Tunnels, enter 2001 as Source port, check Dynamic, click Add, and finally click Open to connect. You can save this back to the session if you like. - Direct the application to use the SOCKS proxy: set up your browser to use SOCKS v4 proxy at host 127.0.0.1 on port 2001. For Firefox, the setting is under Options -> Advanced -> Network -> Connection Settings. For Chrome, it is under Settings -> Show advanced settings -> Network -> Change proxy settings. You can also use add-ons such as FoxyProxy to easily switch between different proxy settings.
- Check your IP address at, e.g. https://ipleak.net. It should show the IP address of the remote machine now.
SSH-Agent/Keychain/Pageant
To avoid having to enter a password every time you SSH, you can use ssh-agent, pageant from PuTTY or gpg-agent.
- First generate a key pair and copy the public key to the target remote system. Make sure to provide a passphrase.
ssh-keygen -t rsa -b 2048 -f ~/.ssh/id_rsa
scp ~/.ssh/id_rsa.pub user@remote.system:.
ssh user@remote.system 'cat id_rsa.pub >> .ssh/authorized_keys && rm id_rsa.pub'
On Windows, simply use PuttyGen from PuTTY to generate a SSH-2 RSA key.
- Do the following every time you begin a new terminal session:
eval $(ssh-agent)
ssh-add ~/.ssh/id_rsa
This will export a few environment variables so that tools such as ssh know where to request keys.
On Windows, use Pageant to serve keys. Many other software tools (i.e., WinSCP) know how to use PuTTY keys too. You can also create a shortcut to launch PuTTY immediately after adding a key:
C:\PuTTY\pageant.exe C:\path\to\id_rsa.ppk -c C:\PuTTY\putty.exe
- A nuisance with the approach so far is that the relevant environment variables are not preserved between shell sessions. As a result, you have to start many ssh-agent instances. To locate existing ssh-agent sessions (and start new ones if none is found), use keychain (a single-file shell script). If it is not provided by your package manager, simply download and unzip it somewhere. Then do the following or add them to
~/.profile:
eval $(/path/to/keychain --eval ~/.ssh/id_rsa)
eval $(/mnt/c/path/to/weasel-pageant/weasel-pageant -r) # for weasel-pageant
eval $(ssh-pageant -r -a "/tmp/.ssh-pageant-$USER") # for ssh-pageant
This solution replaces ssh-agent + keychain, and will require Pageant to be running.
GPG-Agent
GnuPG is an encryption and digital signing software package used for emails (i.e., Mailvelope or Mymail-Crypt for GMail, and EnigMail for Thunderbird), signing code, etc. It provides its own key daemon which can be a drop-in replacement for ssh-agent + keychain and Pageant.
- Useful common settings in
gpg-agent.conf
The following specifies that a passphrase is remembered for 57600 seconds (16 hours) from its last usage so you don’t need to enter it again if you are actively working and making connections. The passphrase will however expire at least once every 999999 seconds (~ 11.6 days).
default-cache-ttl 57600
default-cache-ttl-ssh 57600
max-cache-ttl 999999
max-cache-ttl-ssh 999999
sudo apt install gnupg-agent dbus-user-session
echo enable-ssh-support >> ~/.gnupg/gpg-agent.conf
and comment out “use-ssh-agent” from /etc/X11/Xsession.options to disable ssh-agent.
Then start gpg-agent and import existing SSH keys via ssh-add:
gpg-agent --daemon
ssh-add ~/.ssh/id_rsa
It will first ask you for the passphrase to decrypt the private key, and immediately ask you again for a passphrase to encrypt and store it with gpg-agent. This step needs to be run only once, and gpg-agent will remember the authentication keys across sessions. In addition, gpg-agent itself will be started automatically due to /etc/X11/Xsession.d/90gpg-agent. When a program needs to authenticate, gpg-agent will prompt you for the passphrase if the key has not been cached.
brew install gpg pinentry-mac
echo 'pinentry-program /usr/local/bin/pinentry-mac' >> ~/.gnupg/gpg-agent.conf
echo enable-ssh-support >> ~/.gnupg/gpg-agent.conf
then start gpg-agent and import existing keys as above. Unlike in Linux, the environmental variables are not set up automatically, so you need to do or add the following to ~/.profile
export SSH_AUTH_SOCK="$(gpgconf --list-dirs agent-ssh-socket)"
- On Windows, use the installer provided by GnuPG, and add
enable-putty-support to %AppData%\gnupg\gpg-agent.conf. You can create new authentication-only keys for gpg-agent, but it is not clear how to import existing SSH keys (like we can do with ssh-add under Linux or MacOS). You can, however, copy ~/.gnupg/sshcontrol and relevant keys from .gnupg/private-keys-v1.d from your Linux/Mac machine to %AppData%\gnupg. Optionally, you can create a shortcut to start gpg-agent:
"%ProgramFiles(x86)%\GnuPG\bin\gpg-connect-agent.exe" /bye
PuTTY, WinSCP and other programs should then be able to authenticate as usual.
Customizing .ssh/config
Sharing sessions over a single connection
Add the following to the end of ~/.ssh/config:
Host *
ServerAliveInterval 300
ServerAliveCountMax 3
TCPKeepAlive no
ControlMaster auto
ControlPath ~/mux-%L-%r@%h:%p
ControlPersist 30m
Host alias
If you would like to be able to ssh hostA instead of ssh userX@hostA.dept.univ.edu, add the following to ~/.ssh/config:
Host hostA
HostName hostA.dept.univ.edu
User userX
Multi-hop
- If you need to SSH to a machine via a proxy node, you can do:
ssh -t user1@proxy.system ssh user2@remote.system
This quickly becomes cumbersome, especially if you use scp or rsync too. Automate it in ~/.ssh/config:
Host mesabi ln000? cn0???
HostName remote.system
User user2
ProxyCommand ssh user1@proxy.system nc %h %p
You can then simply do: ssh mesabi.
- Logging into some HPC systems (e.g.,
very.secure.system) requires a hardware token in addition to password. To avoid doing it every time, you can first set up ~/.ssh/authorized_keys in very.secure.system, and SSH to it on a different, long-running computer:
ssh -L1322:localhost:22 -fN user@very.secure.system
This step needs the hardware token. Now on other computers you use, add to ~/.ssh/config:
Host cascade glogin*
HostName localhost
Port 1322
User user
ProxyCommand ssh user2@long.running.system nc %h %p
after which you can ssh cascade without needing the hardware token. It will connect to very.secure.system via port 1322 at long.running.system. However, ALCF’s machines have disabled authentication by SSH keys, so this method will not work there.
Remote Desktop
If you want remote desktop functionality, there are a few options. NoMachine and TeamViewer are so fast that you can even stream HD videos over Internet.
- VNC is a cross-platform solution. A server program needs to be run on the remote end, and a viewer program is used to connect to the server. Mac OS X has built-in VNC server support (System Preferences -> Sharing -> Screening Sharing), but it is not as fast as other implementations (such as RealVNC, TightVNC, and UltraVNC).
- NoMachine started as an improvement of the X Window System by compressing and transferring only the changed region of the screen and wrapping the connections in SSH for encryption. Version 4 began to provide remote desktop between machines that have both installed the software, in addition to connecting to NoMachine servers that are specifically set up. Open-source server implementations(FreeNX, Neatx) exist for Linux. MSI and College of Science and Engineering Labs provides NX servers, from which you can connect to other HPC machines and your workstation. The sessions are preserved unless you terminate them, so you will not lose any ongoing work because of dropped connections and can pick up where you were when you go back and forth between office and home.
- TeamViewer supports remote control, online meeting, and VPN. It runs on Microsoft Windows, Mac OS X, Linux, Android, iOS, Windows RT, and Windows Phone.
